Beam Angle Optimization for Radiative Cancer Therapy#
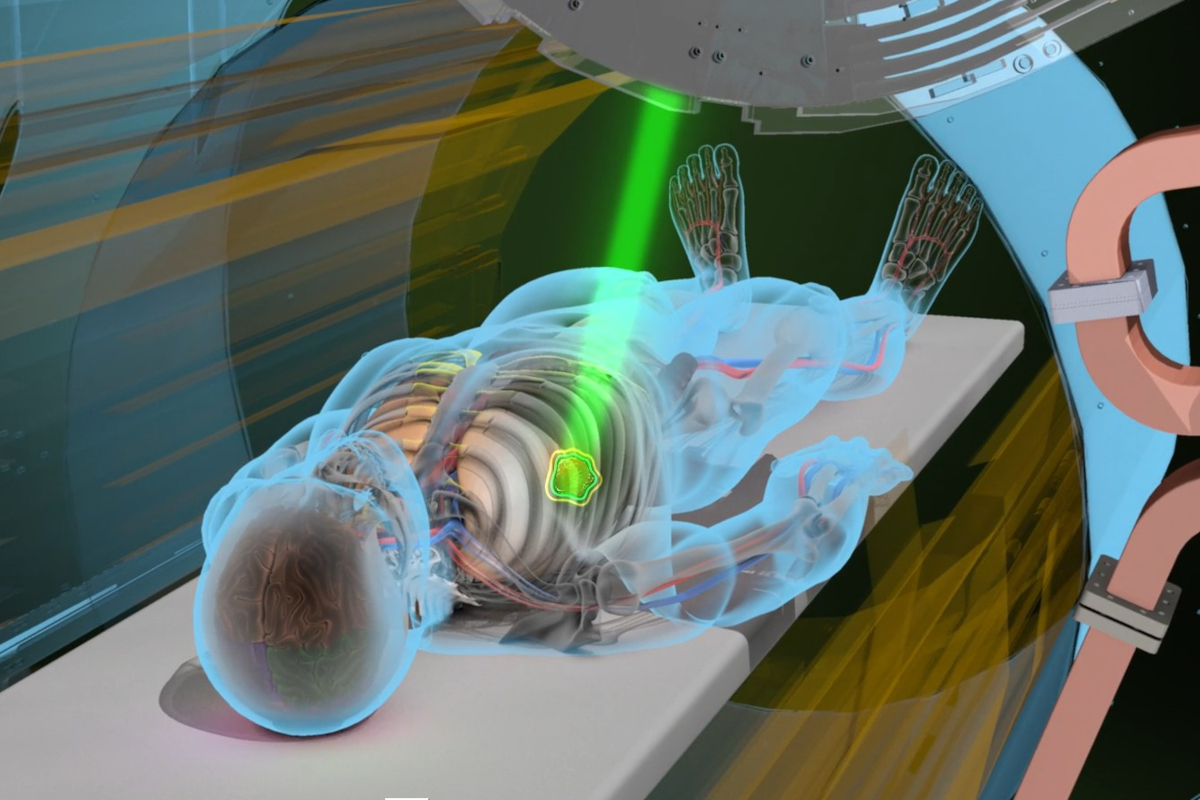
Overview#
In November 1895, Wilhelm Röntgen accidentally discovered X-rays while fiddling with cathode ray tubes. He proceeded to pass these rays through his wife’s hand to create the world’s first X-ray image. This was a pivotal time for the field of medical physics (and their marriage): a means to harness physics phenomena to better the human condition.
Today, radiation therapy involves firing intense beams of ionizing radiation to obliterate cancer cell DNA and halt the malicious growth of tumors. A patient is often laid before a gantry, essentially a LINAC accelerator, that ejects X-rays (or protons, when further precision is required). A rigorous treatment plan requires consideration of:
The ‘dose’ of radiation that a patient can handle (determined by understanding the physical interactions of radiation with tissue)
Beam-Angle Optimization (BAO): annihilating cancerous tissue while preserving healthy cells. BAO involves finding the ideal set of beam paths to accomplish precisely this goal. The machine learning methods we’ve studied thus far have been instrumental in better resolving this combinatorial problem
We will be extracting patient head-neck CT scan + mask data (our ‘input’) from the medical physics OpenKBP challenge. This is real clinical data, and you could make significant contributions to healthcare and cancer treatment by going above & beyond what you learn here!
Data Sources#
Original Source
Local Copy URL
Questions#
Question 01: Absorption & Scattering#
X-rays attenuate exponentially upon entering the body by the Beer-Lambert Law.
What physical phenomena govern this loss of intensity (state at least 3 specific processes)? Which one dominates radiotherapy (which uses ~6 MeV photons)? It’s easy to conclude that the highest dose of radiation is delivered at the skin, why is this untrue?
Question 02: Dosimetry#
We quantify the impact of radiation on tissue (be it healthy or cancerous) via a metric known as Dose. It is the ratio between the energy absorbed and the mass of the tissue absorbing it: measured in Gray (Gy).
What are the implications of too little or too much Dose? Does the type of body tissue matter? Putting it all together, what are the factors which determine the ideal beam angles a gantry should use? For instance, what are the risks to patient health? Why do these considerations make Beam Angle Optimization (BAO) an NP-hard problem?
Question 03: Proton Therapy#
If we used proton therapy instead of X-rays, how would our considerations evolve?
Question 04: Preparing Tensors#
Run the cell below and unzip the contents of openkbp_patient_data into a folder. It should be present in Colab’s (/content/..) or local directory. Note that you will need to adjust base_path to lead to this folder.
Each patient hosts the following three files:
ct.csv - 3D grayscale CT scan of patient anatomy composed of 2D slices. Each voxel represents how much X-ray is absorbed by the tissue in Hounsfield Units (HU)
PTV63.csv - Planning Target Volume (PTV) is a sparse mask that maps the location of the tumor. We’d like binary mask so that 1 is tumor, 0 is no-tumor.
The following are sparse masks of Organs at Risk (OAR), representing what we’d like our beams to avoid
SpinalCord.csv- Irradiating the spine can cause a serious condition known as radiation myelopathy.Brainstem.csv- Radiation causes a vast myriad of devastating effects like RIBN, Ataxia, or even comas.LeftParotid.csv,RightParotid.csv- Irradiating salivary glands can cause xerostomia (dry mouth) or worseMandible.csv- The mandible has a limited blood supply, and osteoradionecrosis can occur YEARS after treatment.
These are flattened volumes with voxel indices. The cell below converts the csv files into binary 3D volumes [128, 128, 128] where each point represents an intensity (for the ct scan) or binary value (for ptv & spinal cord). You will have an optional dictionary of patient_data to access for your convenience.
Your tasks:
Add a line to the for loop that stacks
ct,ptv, andoars(Organs at Risk) into a single tensor. Note that oars is a dictionary you may need to unpack. What’s the final shape of this tensor? Draw an analogy to an RGB image.Visualize a single 2D slice (axial, sagittal, or coronal) of a patient’s CT scan with the tumor(ptv) overlaid…as a sanity check.
!wget https://github.com/florilegium7/Physics-informed-DQN-Radiotherapy/releases/download/v1/openkbp_patient_data.zip
!unzip openkbp_patient_data.zip -d openkbp_patient_data
import numpy as np
import pandas as pd
import os
base_path = 'openkbp_patient_data' #you may adjust this (e.g. /content/openkbp_patient_data)
patient_ids = ["pt_1", "pt_2", "pt_9","pt_40","pt_68","pt_70", "pt_90", "pt_91" , "pt_99", "pt_187"]
patient_tensors = []
def mask_from_sparse(path,shape=(128, 128,128)): #converts voxel indices to 3D binary masks!
indices = pd.read_csv(path, header=None)[0].dropna().astype(int).values
mask_flat = np.zeros(np.prod(shape), dtype=np.uint8)
mask_flat[indices] = 1
return mask_flat.reshape(shape)
def sparse_to_ct_volume(path, shape=(128, 128, 128)): #turns the csv into full 3D volume CT scans
df = pd.read_csv(path, header=None).dropna()
indices = df[0].astype(int).values
values = df[1].astype(float).values
ct_flat = np.zeros(np.prod(shape), dtype=np.float32)
ct_flat[indices] = values
return ct_flat.reshape(shape)
#128 x 128 x 128 pixels : (depth, height, width)
for pt_id in patient_ids:
pt_dir = os.path.join(base_path, pt_id)
ct = sparse_to_ct_volume(os.path.join(pt_dir, "ct.csv"))
ptv = mask_from_sparse(os.path.join(pt_dir, "PTV63.csv"))
oars = {} #dictionary
organs = ["SpinalCord", "Brainstem", "LeftParotid", "RightParotid", "Mandible"]
for organ in organs:
organ_path = os.path.join(pt_dir, f"{organ}.csv")
if os.path.exists(organ_path):
oars[organ] = mask_from_sparse(organ_path)
patient_data = {
"id": pt_id,
"ct": ct, #ct scan -> 3D volume with intensities
"ptv": ptv, #tumor -> 3D binary mask (1 means tumor!)
"oars": oars #dictionary of organs at risk -> each is a 3D binary mask
}
pt_tensor = #Your Code Here
patient_tensors.append(pt_tensor)
#Your slice visualization here:
import matplotlib.pyplot as plt
Question 05: Reinforcement Learning#
To simulate the effects of a naive beam (simple straight lines) passing through the patient, we provide a ‘beam mask’ that creates a beam onto a 2D slice of the patient scan. We assume there are 36 possible angles from which a beam can be fired (one every 10 degrees). dose is a 2D heatmap that represents how much radiation is delivered to each pixel in your slice after the passage of many beams.
Create a Reinforcement Learning Model which selects beam angles that provide radiation dosage to the tumor while avoiding critical organs.
Populate the training loop by:
Creating a policy that determines the next
angleCreating a Q-learning update
Define a reward function that heavily penalizes a beam that passes through an organ and rewards a beam that hits the tumor/ptv (the output should be a score)
Tweak parameters if you so desire
Print the top 3 beam angles after training is complete
import random
beam_angles = np.arange(0, 360, 10) #[0, 10, 20, ..., 350]
max_beams = 5 #ensures beams are chosen strategically
episodes = 500
alpha = 0.1 #learning rate
epsilon = 0.2 #exploration rate
slice_shape = (128,128) #a single 2D slice
from skimage.draw import line_nd
def generate_beam(angle_deg, shape): #beam mask generator!
h, w = shape
beam_mask = np.zeros((h, w), dtype=np.uint8)
center = np.array([h // 2, w // 2])
length = max(h, w)
angle_rad = np.deg2rad(angle_deg)
dx = np.cos(angle_rad)
dy = np.sin(angle_rad)
start = (center - length * np.array([dy, dx])).astype(int)
end = (center + length * np.array([dy, dx])).astype(int)
start = np.clip(start, 0, [h - 1, w - 1])
end = np.clip(end, 0, [h - 1, w - 1])
rr, cc = line_nd(start, end)
beam_mask[rr, cc] = 1
return beam_mask
for episode in range(episodes):
#Randomly select a patient from patient_tensors HERE
ct,ptv =
#extracts a single 2D slice from the middle
mid = ct.shape[2] // 2
ct_slice = ct[:, :, mid]
ptv_slice = ptv[:, :, mid]
#extract organ slices HERE
dose = np.zeros(slice_shape, dtype=np.float32)
selected_angles = []
for i in range(max_beams):
#Your policy here
beam = generate_beam(angle, slice_shape)
dose += beam.astype(np.float32) #adds 'radiation' to the pixels
selected_angles.append(angle)
reward_score = reward(dose, ptv, organs)
for angle in selected_angles:
#Q-learning update here
def reward(dose, ptv, organs): #your reward function HERE
raise NotImplementedError()
Q = np.zeros(len(beam_angles)) #Q-table
Your model (have you named it yet?) has chosen its ideal beam angles, let’s cross our fingers, fire these into our patient, and see what we get!
patient = random.choice(patient_tensors)
mid = patient.shape[3] // 2
ct, ptv = patient[0, :, :, mid], patient[1, :, :, mid]
shape = ct.shape
dose = np.zeros(shape, dtype=np.float32)
top_indices = sorted(range(len(Q)), key=lambda i: -Q[i])[:max_beams]
selected_angles = [beam_angles[i] for i in top_indices]
for angle in selected_angles:
beam = generate_beam(angle, shape)
dose += beam.astype(np.float32)
plt.imshow(ct, cmap='gray')
plt.imshow(dose, cmap='hot', alpha=0.4)
plt.contour(ptv, colors='green')
plt.contour(cord, colors='blue')
for angle in selected_angles:
plt.imshow(generate_beam(angle, shape), cmap='Reds', alpha=0.2)
plt.title(f"Selected Beams: {selected_angles}\nPTV: Green, Cord: Blue")
plt.axis('off')
plt.show()
Question 06: Physics-informed reinforcement learning#
Ready to boogie? We’re about to imbue physics into our RL model. Let’s start by making our beam of ionizing radiation more realistic using what we discussed in Question 01. Adjust the decay in generate_true_beam below so that it actually attenuates our beam (energy loss as it passes through tissue)
from scipy.ndimage import center_of_mass
def generate_physical_beam(angle_deg, ptv_mask, shape = (128,128), spread_sigma=2.0, attenuation_coeff=0.01):
h, w = shape
beam_mask = np.zeros((h, w), dtype=np.float32)
if isinstance(ptv_mask, torch.Tensor):
ptv_mask_np = ptv_mask.detach().cpu().numpy()
else:
ptv_mask_np = ptv_mask
tumor_center = np.array(center_of_mass(ptv_mask_np))
center = tumor_center.astype(int)
length = max(h, w)
angle_rad = np.deg2rad(angle_deg)
dx = np.cos(angle_rad)
dy = np.sin(angle_rad)
start = (center - length * np.array([dy, dx])).astype(int)
end = (center + length * np.array([dy, dx])).astype(int)
start = np.clip(start, 0, [h - 1, w - 1])
end = np.clip(end, 0, [h - 1, w - 1])
rr, cc = line_nd(start, end)
rr = np.clip(rr, 0, h-1)
cc = np.clip(cc, 0, w-1)
for i, (r, c) in enumerate(zip(rr, cc)):
decay = 1*np.exp(-attenuation_coeff*i)
beam_mask[r, c] += decay
for dr in range(-3, 4): #gaussian spread via scattering
for dc in range(-3, 4):
r_spread = r + dr
c_spread = c + dc
if 0 <= r_spread < h and 0 <= c_spread < w:
distance = np.sqrt(dr**2 + dc**2)
spread_value = np.exp(- (distance**2) / (2 * spread_sigma**2))
beam_mask[r_spread, c_spread] += decay * spread_value
beam_mask = np.clip(beam_mask, 0, 1.0)
return beam_mask
We’d like to inject a physics loss that encodes the actual attenuation and radiation dose delivery of the beam, rewarding correct dose distribution between the spine and tumor. We need to upgrade our simple Q-learning method into a Deep-Q Network (DQN). This should only require simple structural changes (refer to our reinforcement learning notebook). Typically, these would use a Bellman loss. With our new physics-loss it should resemble: $\( L_{\text{Total}} = L_{\text{Bellman}} + \lambda_{\text{Physics}} L_{\text{Physics}} \)$
The physics loss takes the following form: $\( L_{\text{Physics}} = \lambda_{\text{PTV}} \cdot \text{Underdose}_{\text{PTV}} + \lambda_{\text{Spine}} \cdot \text{Overdose}_{\text{Organs}} \)\( Each \)\lambda\( assigns 'weight' to a term, tinkering with these may prove beneficial. The Underdose term penalizes not giving the tumor the maximum dose (1) \)\( \text{Underdose}_{\text{PTV}} = \mathbb{E} \left[ \max \left( 0,\ 1 - D_{\text{PTV}} \right) \right] \)$
\(D_{\text{PTV}}\): dose received by pixels inside the tumor (PTV mask)
\(1 - D_{\text{PTV}}\): underdose per pixel (1 is max dose)
\(\max(0, 1 - D_{\text{PTV}})\): penalizes only underdosed regions
\(\mathbb{E}[\cdot]\): average
The Overdose term penalizes giving the organs too much dose $\( \text{Overdose}_{\text{Organs}} = \mathbb{E} \left[ \max \left( 0,\ D_{\text{Organs}} - D_{\text{safe}} \right) \right] \)$
\(D_{\text{safe}}\): Safety threshold for organ (max dose it can receive). You can find these online
Now create that network, like a true ML-infused medical physicist of the 21st century would!
#Your masterpiece here
For our grand finale we will overlay your newfound results onto a CT scan of a tumor. Congratulations! You just made a treatment plan for a cancer patient!!
from matplotlib.lines import Line2D
def visualize_beams_on_ct(patient, selected_angles, slice_index= None, shape = (128,128)):
channel, height, width, depth = patient.shape
if slice_index is None:
slice_index = depth // 2 #default to middle slice
ct_slice = patient[0, :, :, slice_index]
ptv_slice = patient[1, :, :, slice_index]
organ_slices = patient[2:, :, :, slice_index]
dose = np.zeros(shape, dtype=np.float32)
for angle in selected_angles:
beam = generate_physical_beam(angle, ptv_slice, shape)
dose += beam.astype(np.float32)
dose_norm = dose / np.max(dose + 1e-5)
plt.figure(figsize=(6, 6))
plt.imshow(ct_slice, cmap='gray', alpha=0.6)
plt.imshow(dose_norm, cmap='hot', alpha=0.4) #beam
plt.contour(ptv_slice, colors='red', linewidths=1, label='PTV')
for organ_slice in organ_slices:
plt.contour(organ_slice, colors='blue', linewidths=1)
plt.title("Radiotherapy Treatment Plan")
plt.axis('off')
legend_elements = [
Line2D([0], [0], color='red', lw=2, label='Tumor'),
Line2D([0], [0], color='blue', lw=2, label='Organs at risk')
]
plt.legend(handles=legend_elements, loc='upper right')
plt.show()
(Optional adventures)
If you’d like to spice things up and maybe even contribute to current medical physics research, here’s some fun ideas:
The model & beam currently work on 2D slices, give it 3D awareness!
Imbibe more physics into the beam (e.g. consider penalizing the beam for deviating from real scattering physics in your reward function)
Incorporate consideration of the varying physical properties of tissue for each organ/bone/tumor region
Try finding datasets of real beam angles used in clinical settings and use a supervised learning approach to compare results.
Add more patients from OpenKBP and train for a long, long time. Got any supercomputers handy?
Thanks for doing my project! Feel free to reach out with any questions/comments on the notebook or beyond!! :)
References#
[1] A. Babier, B. Zhang, R. Mahmood, K.L. Moore, T.G. Purdie, A.L. McNiven, T.C.Y. Chan, “OpenKBP: The open-access knowledge-based planning grand challenge and dataset,” Medical Physics, Vol. 48, pp. 5549-5561, 2021.
Acknowledgements#
Initial version: Aarya Mehta with some guidance from Mark Neubauer
© Copyright 2025

